Oh my God, they killed Kenny!
maintenance
Storage system failure
The last reboot of machines (14/12) had revealed a serious problem in our storage system. Long story short, a large part of Fink data could not be recovered, making difficult to get some services back online, and we had no backup for this. It is yet unclear what happened exactly, but most likely a manipulation of the journal of transations in November that silently corrupted the system, only revealed a month later when the systems were rebooted. It was too late unfortunately, with more than 170k transactions missing…
Only the database part was affected (serving the Science Portal), and the time-critical part (that processes nightly data) remained unaffected. Luckily though, the ZTF survey was shut down at the same moment due to out of order cryocooler – so there were no alerts sent during the Fink shutdown.
We took the opportunity of this maintenance to learn lessons from the post-mortem analysis, refine the roadmap for the infrastructure, consolidate the weak points, and deploy new services that were on tests. We describe below the epic recovery that took place during the 2021 Christmas break to bring back all Fink data from scratch.
Summary for the recovery
For those who do not want to read the long blabla below, here are the salient results.
The reprocessing went well. We restored the storage system, and we made it more robust to prevent similar failures. We optimised some parts of the pipelines, and took the opportunity of digging into old data to retrieve more data than before (nearly doubled!). The timings for the reprocessing were:
| Step | Task | Throughput per task | Time for 4.7 TB |
|---|---|---|---|
| Download | 5 nights | 300 MB/s | 5.5 hours |
| Decompress+merge | 1 night | 33 MB/s | 42 hours |
| Upload to HDFS | 16 nights | 300 MB/s | 5.5 hours |
| Science reprocessing | 1 night | 15 MB/s | 96 hours |
So we could recreate everything for 2020 and 2021 in about one week. Note that with a wiser parallelisation of the decompression step, we could reach +100 MB/s, that is a step time divided by more than 2. Note that also the science reprocessing step could be speed-up by increasing the size our HBase instance.
In terms of available alerts, the number has roughly doubled (11/2019 to 12/2021):
| Previously | Current | |
|---|---|---|
| Total number of alerts processed | 41,370,359 | 72,017,228 |
| Classified | 17,189,220 | 35,946,715 |
| Unclassified | 24,181,139 | 36,070,513 |
Note that there are more alerts classified (in percentage) than before. This is due to 2 main reasons:
- With more data, classifiers get better, and more alerts are tagged.
- CDS SIMBAD database get updated continuously – so alerts from previous years benefit from the most recent SIMBAD entries, and many previously unknown alerts are now tagged.
Resurrecting HDFS
HDFS was severly corrupted. The journal of transactions was full of gaps (>170k missing transactions), and despite several attempts to save it, we could not restart our databases. Long story short, HDFS is made of blocks of data. We found that many blocks were affected, and we could just remove those blocks (which would mean losing 1 month of data), and starting again from this point. But not only newly created blocks were affected during the troubled times, but also previously created blocks that were updated. The 2nd case is typical for the database, managed by HBase. Basically, nearly all blocks for the database were updated at some point, and it was impossible to restart HBase (blocks with metadata were corrupted and unavailable).
So after a long reflection, we came to the conclusion that despite all our efforts, it was just the moment to reformat HDFS. But we also put in place several mechanisms to prevent future similar failures:
- Installation of a second NameNode (Standby NameNode) to take over in case of failure.
- Regular back-up of the critical data.
- Regular audits of the infrastructure (to avoid long drifts of unusual situations like we had).
Re-building the raw data lake
Downloading & formatting raw ZTF data
Public ZTF alert data can be downloaded from https://ztf.uw.edu/alerts/public. For 2021 & 2020, this corresponds to roughly 4.7 TB of compressed data. We used aria2c to speed-up the download, and we managed to get a rather stable transfer at 60 MB/s per file, for 5 files simultaneously (i.e. 300 MB/s). The transfer of 4.7 TB took about 5.5 hours:
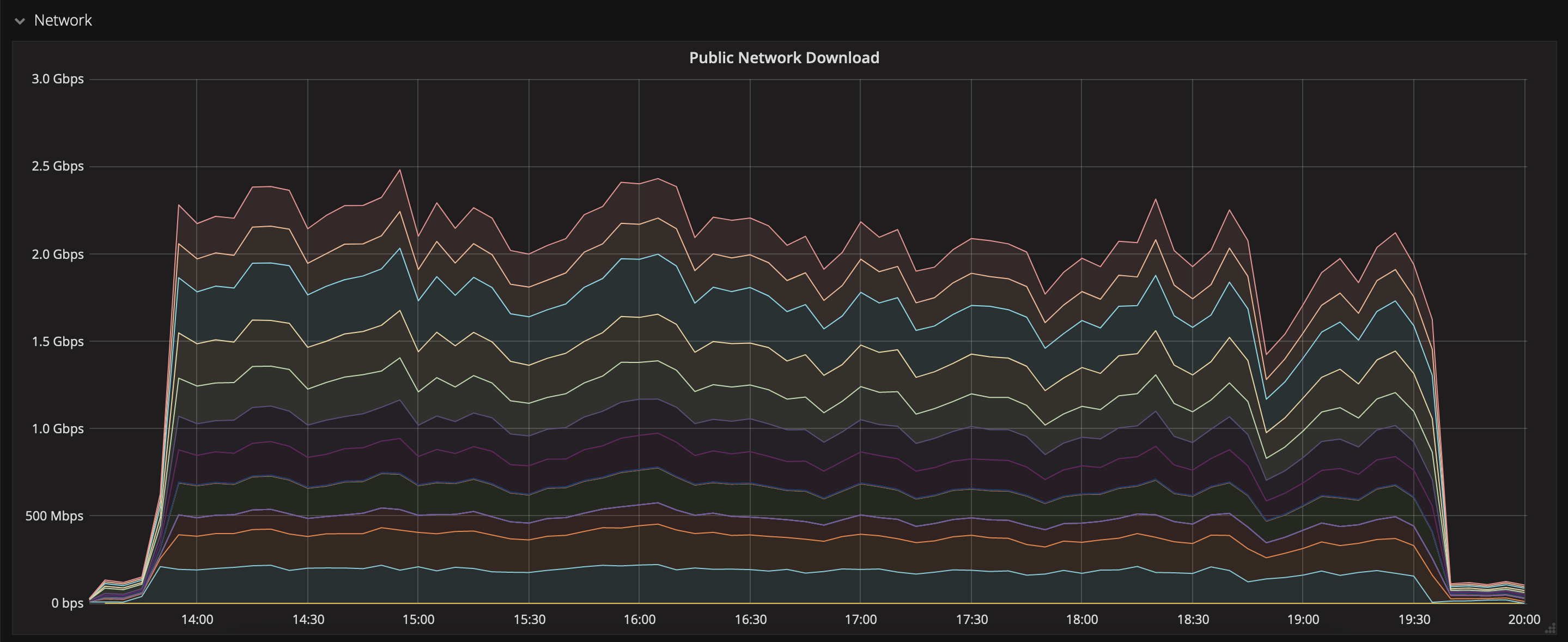
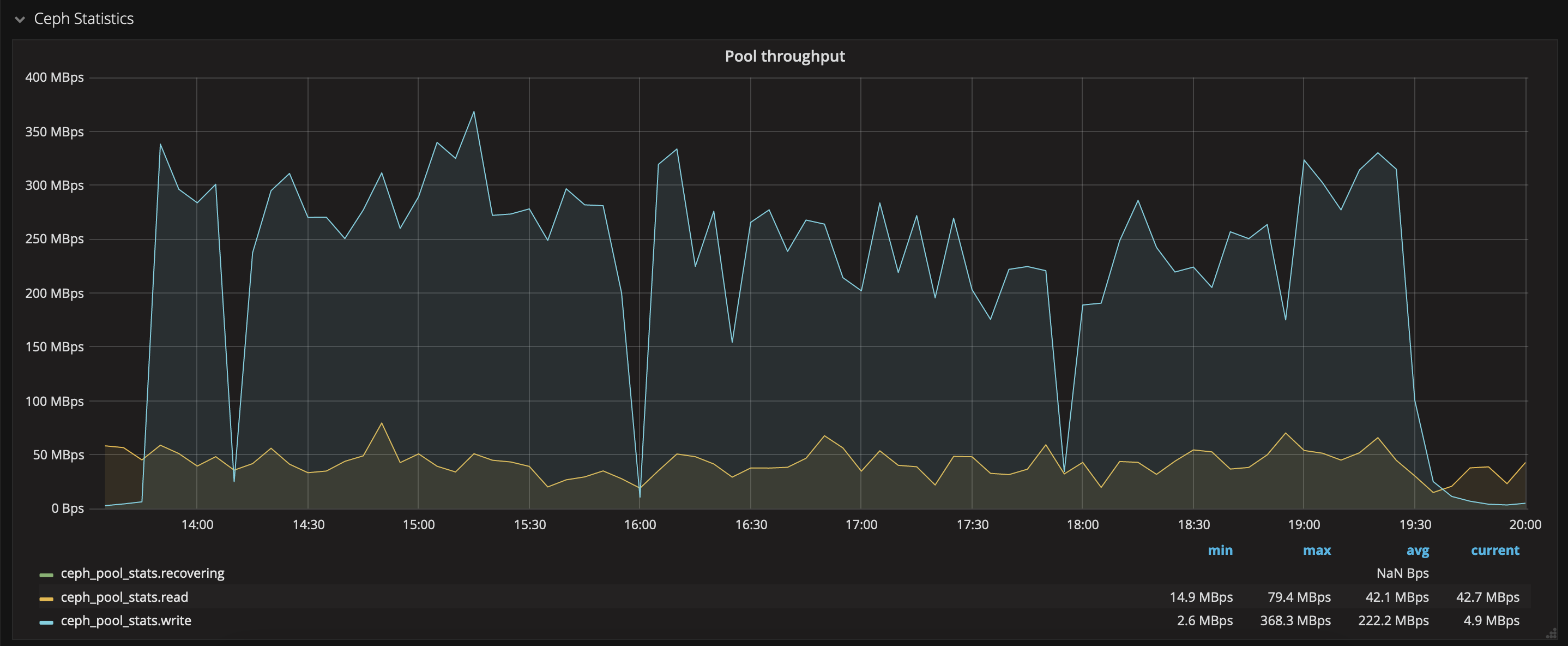
and the writing on CEPH was quite stable as well:
Format files for Apache Spark
The data transfered is made of compressed folders (.tar.gz) containing Apache Avro files. While this is good for an archive, this is not suitable for our processing. You need then to decompress the files, concatenate the data from individual Avro files into larger chunks, and save into Parquet files.
The decompression is the slowest operation, and it is difficult to parallelize inside each task. We managed to slightly speed-up the process by using pigz and tar instead of tar alone (factor of 2 faster). The decompression rate was about 80 MB/s per file.
Once data is decompressed, we obtain one Avro file per alert, and there are hundreds of thousands alerts by folder! This deluge of small files is bad for the processing. Hence, the next operation is to concatenate many alerts at once. In addition, we perform a conversion from Avro to Parquet (the chosen format for internal operations in Fink). By concatenating 3000 raw Avro files into one single Parquet file (with snappy compression), we currently achieve a compression factor of about 1.6. Finally, files are saved into a partitioned structure (year/month/day) to speed-up subsequent operations.
The wrapper will look for all ztf_public*.tar.gz files, and do the job. To speed-up the merge operation, we use mpi under the hood. The script will use by default 16 cores – leading to a writing throughput of about 170 MB/s.
As decompression and merge are done one after the other, and file by file, we shall obtain a rate of about 55 MB/s. In practice, we observe twice slower (why?). Note that it would have been perhaps wiser to parallelize the decompression step first (by parallelizing folders), and then parallelize the merging step (that way we would easily reach +300 MB/s for decompression, that is a total rate at +100 MB/s). Improvement for the future!
Reprocessing science data
We then move all the data to our HDFS cluster, using a simple client (https://hdfscli.readthedocs.io/en/latest/). This took roughly 5h for the 4.7 TB. Once the data was in place, we could relaunch the science processing, and populate the databases.
But since we were restarting from scratch, why not improving the pipelines and the data quality? Since the beginning of Fink, we learned a lot, and our approach to the data is different from 2 years ago. We identified two major upgrades (and small ones): quality cuts and science module speed.
Quality cuts
Quality cuts are the entry cuts that select valid alerts, and discard “bad quality” alerts before the processing. Our first set of quality cuts includes:
- RealBogus scores assigned by the ZTF alert distribution pipeline Mahabal et al. (2019); Duev et al. (2019). The values must be above 0.55.
- Number of prior-tagged bad pixels in a 5 x 5 pixel stamp. The value must be 0.
- The difference between the aperture magnitude and the PSF-fit magnitude. The absolute value must be lower than 0.1.
This filtering reduces about 70% of the initial ZTF public stream, leaving 30% of all collected alerts to be further processed by the science modules. After 2 years of ZTF data, we found out that the last cut was not really motivated given our science cases. A lot of alerts were discarded, while they were just “noisy” – but perfectly valid from the science point of view. Hence, in this reprocessing, we only activated the first two cuts.
This change of the cut definition has two effects: (1) it lets more alerts to enter the pipeline (+30% of the total alerts, that is doubling the previous number of alerts), and (2) give more data for the alert classifiers to perform their task (and eventually allow to classify objects earlier, as first alerts are usually noisier than others).
Science module speed-up
Science modules were slow – at least some of them. We managed to get a huge speed-up with respect to current version, by just moving the candidate filtering before the processing instead of after. Of course this gives less flexibility in the alert selection afterwards (since, for each science module, you process only what satisfies the science case cuts), but at least you stop classifying obvious solar system objects inside the supernova module for example…
Overall throughput
The steps from raw data to the HBase tables are:
- load the raw data, filter for valid alerts, run science modules, save on disk.
- load previously stored science data, and push to the main table in HBase.
- load previously stored science data, select data based on needs, and push to the index tables in HBase.
So there are: 1 time processing, 2 times load, 1 time write directly in HDFS, 1 time write in HBase (which uses HDFS), 11 times write in HBase (thin index tables). This is a lot of write… Using 36 cores, we reached a total throughput of 300 alerts/s, that is roughly 10 MB/s… This is super slow! Note that increasing the number of cores does not speed-up the computation for two reasons: (1) there are perfidious join operations in the processing, and (2) our HBase instance is too small to perform heavier write operations. The HBase instance is a single machine with 6 cores, each with 2GB RAM – this is clearly suboptimal given the current volume of data and have rather large regions, and too many regions per regionserver (about 20 GB per region, and +200 regions in the single regionserver). As a consequence, heavy writes causes the instance to be frequently stuck, with crashes, hence we limit the number of parallel mappers.
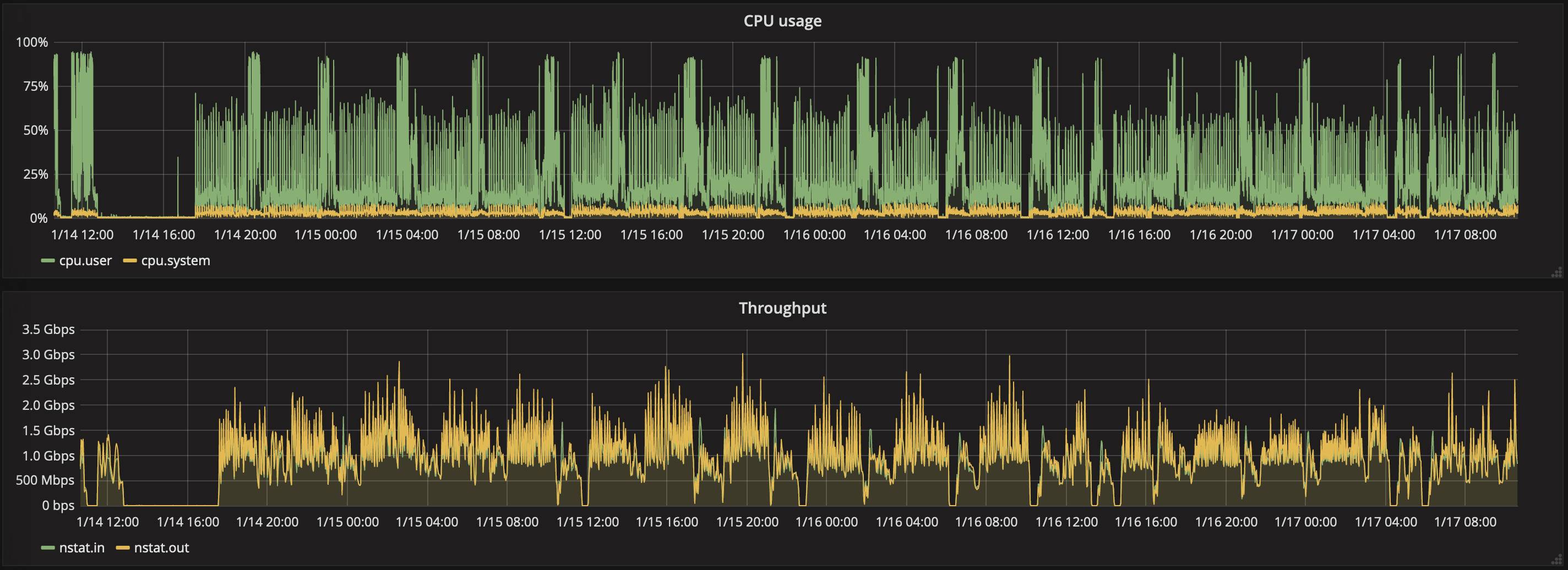 CPU usage & throughput for the HBase instance, during the reprocessing. Note the gaps appearing – they are crash of the instance due to heavy write operations.
CPU usage & throughput for the HBase instance, during the reprocessing. Note the gaps appearing – they are crash of the instance due to heavy write operations.
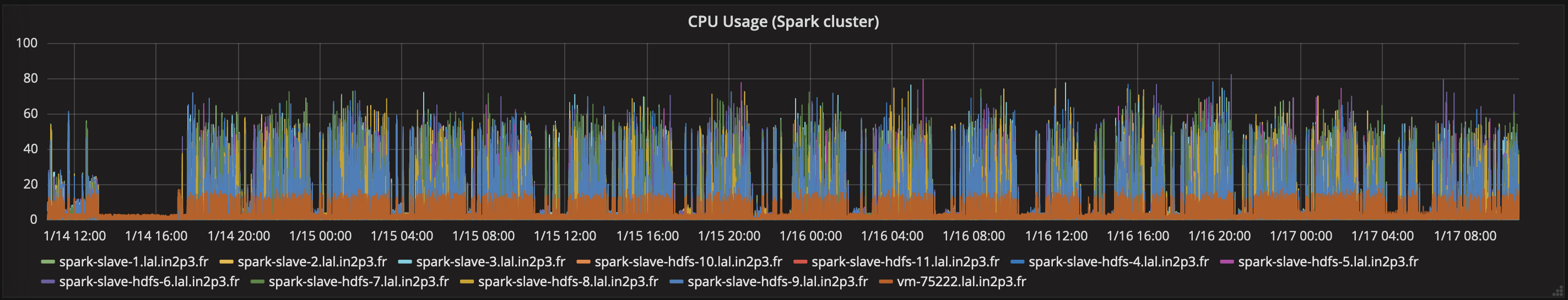 CPU usage for the Apache Spark cluster, during the reprocessing. We used only 36 cores for each task.
CPU usage for the Apache Spark cluster, during the reprocessing. We used only 36 cores for each task.
Changelog
See the full changelog for this maintenance: